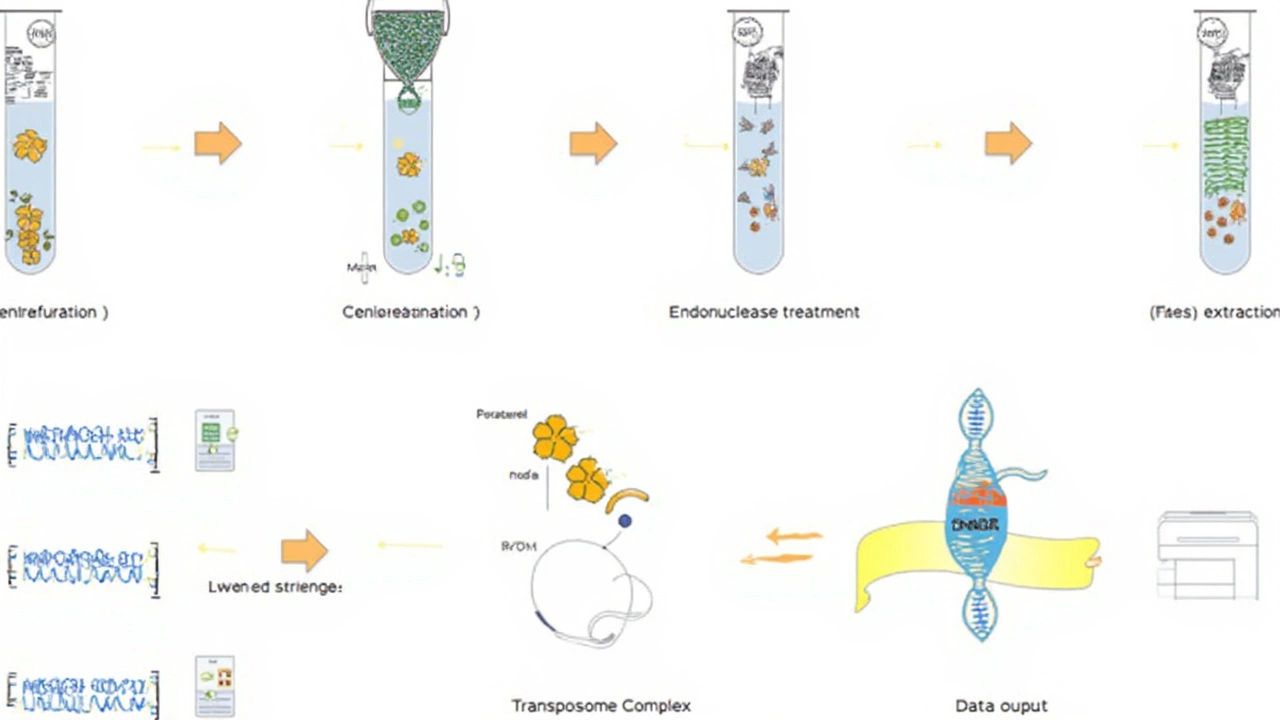
Breaking the Mold: Fast Pathogen Detection With Unified Metagenomics
When someone is sick, waiting days for lab results can feel like an eternity. Now, imagine diagnosing complex infections—in real time, at the patient’s bedside, no less. That’s where a unified metagenomic method using nanopore sequencing comes in, turning the usual wait into just hours for comprehensive answers.
Researchers, including Themoula Charalampous, have developed a workflow that puts traditional multi-assay diagnostics to shame. Their approach scans clinical samples for a whole range of microorganisms—whether those troublemakers have RNA or DNA—using one streamlined process. You get a first look at potential pathogens in half an hour, and within 7 hours, you’re staring at a full microbial breakdown.
The difference starts right with the sample prep. Instead of laboriously removing human DNA, the team goes straight for the microbes by mechanically cracking open human cells. This releases everything lurking inside—bacteria, viruses, you name it—and skips a step that usually eats up time and can risk missing important pathogens.
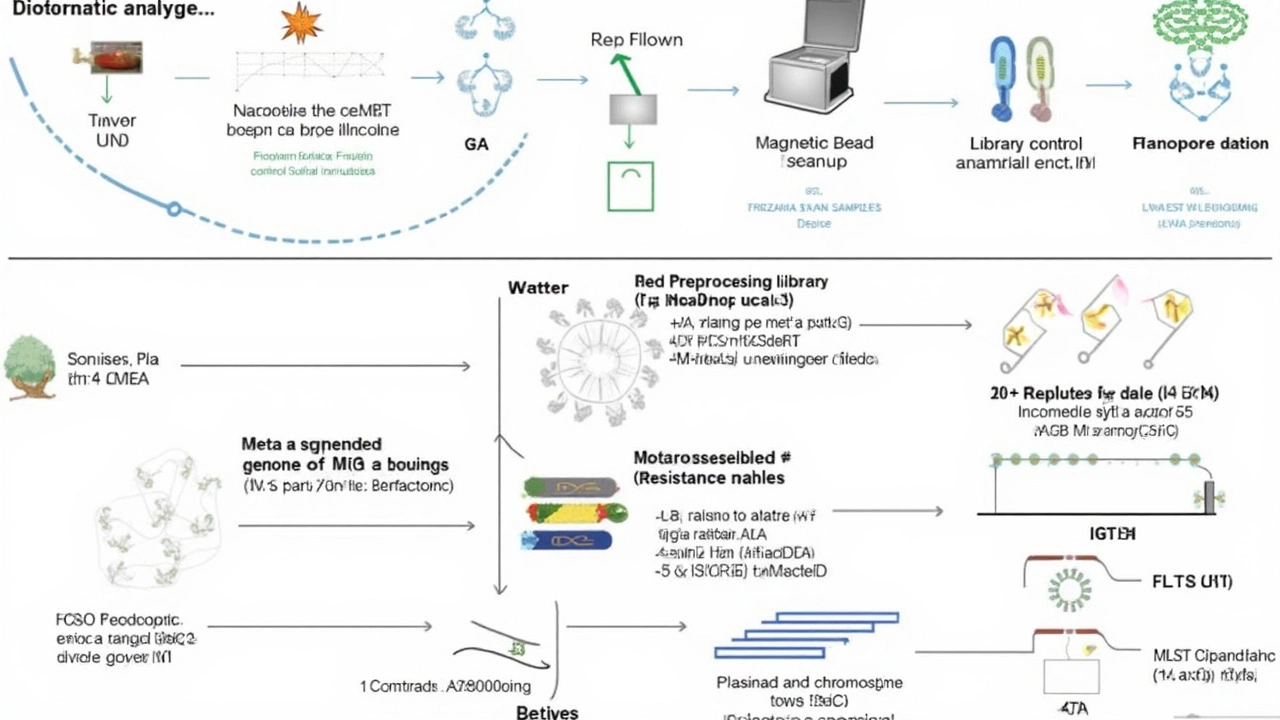
One Workflow, Multiple Pathogens: What Makes It Stand Out?
The real power of this method is its ability to detect both RNA and DNA pathogens in one go. Traditional diagnostics often have you running different tests for different types of bugs—each with their own reagents, steps, and waiting periods. Here, a clinical sample (think sputum or blood) can be fully analyzed using a universal sequencing approach that works on site, even in hospitals or outpatient clinics that aren’t set up with fancy lab infrastructure.
- Nanopore sequencing provides super-fast, real-time data so preliminary results can arrive just 30 minutes after starting.
- The workflow has no need for human DNA depletion, streamlining the process and making it less likely you’ll lose important microbial information.
- Researchers validated the method by testing it on clinical samples suspected of respiratory infection. It accurately identified a wide range of bacteria and viruses from a single sample, giving a much broader picture than old-school targeted tests.
- By offering a full microbial profile, clinicians get more data to guide treatment, fast, and with fewer gaps. This can be a game-changer for things like pneumonia or flu outbreaks, where the usual culture methods can be too slow or even miss mixed infections.
Another kicker? This metagenomic workflow stays sensitive without the elaborate purifying or depletion steps that usually bog down sequencing-based diagnostics. That means you keep the process compact, portable, and surprisingly accessible—all while catching tricky pathogens that sometimes hide in complex samples.
Researchers are excited about its use for respiratory illnesses, but the technology works across different types of infections and sample types. As hospitals keep looking for ways to speed up and sharpen diagnostics (especially during viral seasons or in outbreak scenarios), this new clinical samples tool looks set to shake things up for patients and doctors alike.




